How To Identify Protein Protein Interactions
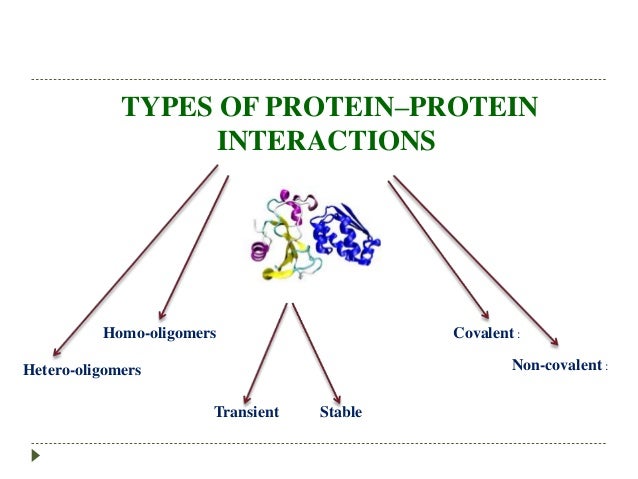
The race to understand cellular function hinges on mapping the intricate web of protein-protein interactions (PPIs). Identifying these interactions is crucial for drug discovery and understanding disease mechanisms.
Scientists are employing diverse techniques to decipher which proteins bind to each other, unveiling potential therapeutic targets and paving the way for personalized medicine. This article highlights the key methods used to detect PPIs, offering a snapshot of the current landscape of proteomic research.
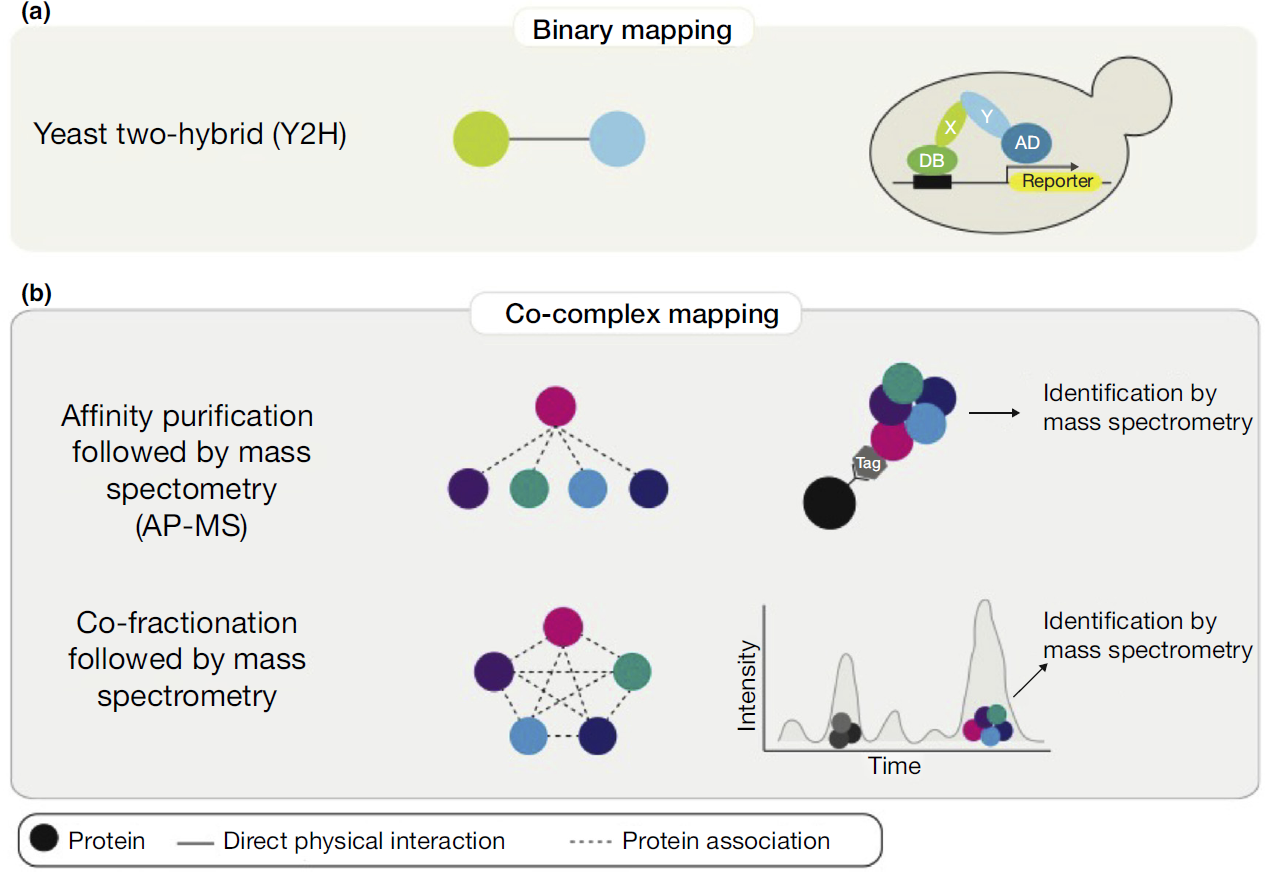
Yeast Two-Hybrid (Y2H) Screening
Y2H remains a widely used method to identify PPIs in vivo. This technique uses genetically engineered yeast cells to detect interactions between a "bait" protein and a "prey" protein.
If the two proteins interact, they activate a reporter gene, signaling a positive interaction. Y2H is known for its ability to screen for interactions across entire proteomes.
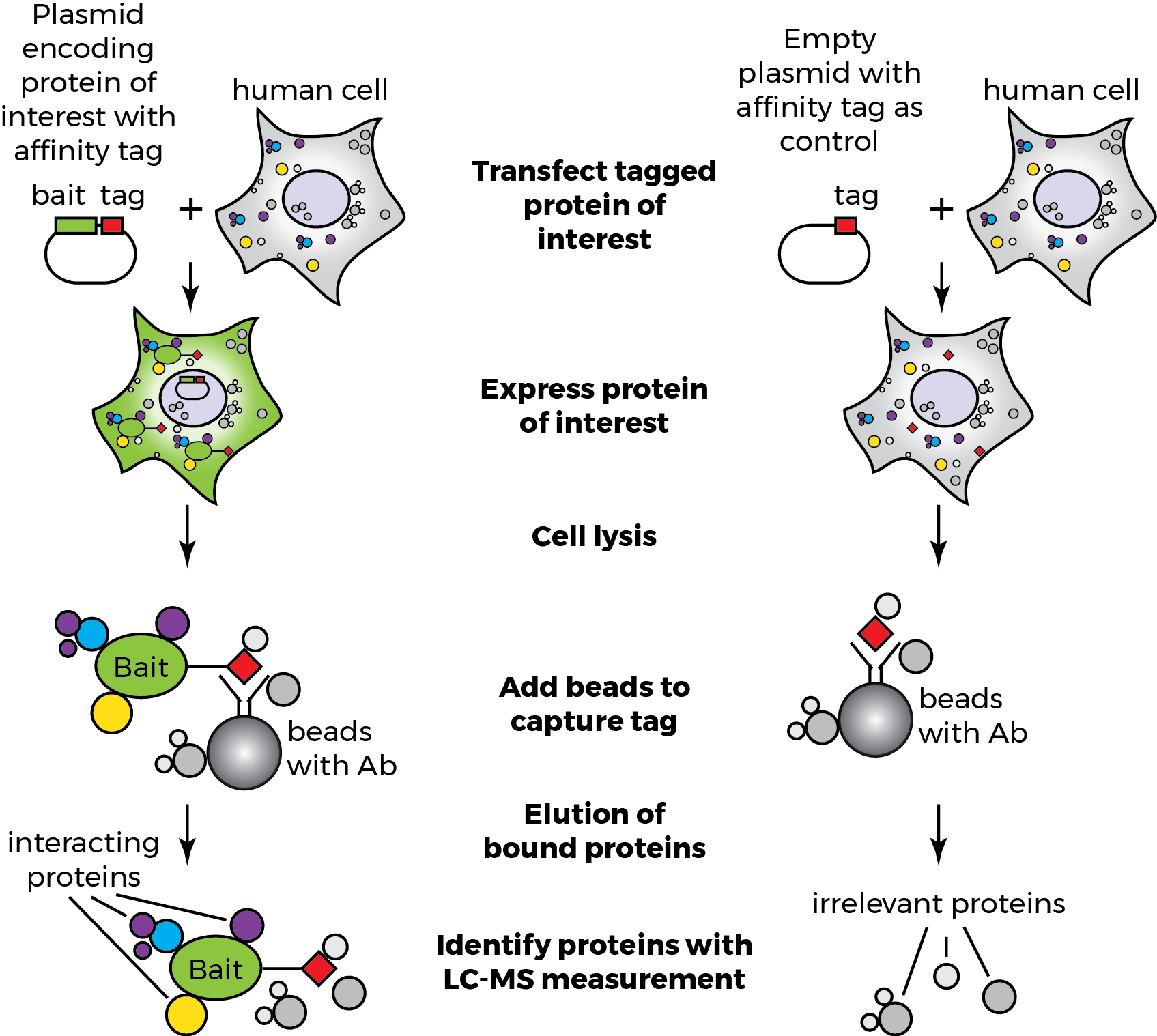
Affinity Purification-Mass Spectrometry (AP-MS)
AP-MS involves isolating a protein complex using an antibody specific to a "bait" protein. The purified complex is then analyzed using mass spectrometry to identify interacting proteins.
This method is highly effective for identifying native protein complexes within a cellular environment. AP-MS offers valuable insights into functional modules within cells.
Surface Plasmon Resonance (SPR)
SPR is a label-free technique that measures changes in refractive index on a sensor surface when molecules interact. This is a powerful biophysical method for studying PPIs in real time.
One protein is immobilized on a sensor chip, and the other protein is passed over the surface. The change in refractive index indicates binding, allowing researchers to quantify the affinity and kinetics of the interaction.
Co-immunoprecipitation (Co-IP)
Co-IP is a classical method for confirming PPIs in vitro or in vivo. This technique involves using an antibody to immunoprecipitate a protein of interest, and then detecting interacting proteins via Western blotting.
Co-IP is frequently used to validate interactions identified by other methods, ensuring the reliability of PPI data. The technique offers a straightforward approach for confirming suspected interactions.
Proximity Ligation Assay (PLA)
PLA allows visualization of PPIs in situ within cells or tissues. Antibodies against two different proteins are used, and if the proteins are in close proximity, a DNA oligo is amplified, producing a fluorescent signal.
This method provides spatial resolution, enabling researchers to study PPIs in specific cellular compartments. PLA allows direct observation of interactions within their native context.
Emerging Technologies and Future Directions
New methods are constantly being developed to improve the accuracy and efficiency of PPI detection. These advancements will be pivotal in understanding complex biological processes.
Cross-linking mass spectrometry (XL-MS) and AlphaFold are revolutionizing structural biology and PPI prediction. They offers detailed structural information about protein complexes, revealing binding interfaces.
Future research will focus on integrating these diverse techniques to generate comprehensive PPI networks. Researchers are working to understand the dynamic nature of protein interactions in various cellular contexts. Unlocking PPIs is paramount for therapeutic development and a deeper understanding of life itself.









-p-2000.png)
